For customers seeking to ensure the safety and efficacy of their CRISPR-based therapeutic developments through off-target analysis, our comprehensive end-to-end services encompass assays from off-target nomination to confirmation, with outputs specifically tailored for IND filings.
"We have been very impressed with the characteristics of Alt-R™ HiFi Cas9 Nuclease, particularly its consistent on-target editing and low off-target activity. We are excited to use this version in our future experiments focused on developing novel genome editing-based therapies for severe diseases with unmet medical needs."

Dr. Matt Porteus
Principal Investigator
Stanford
| Key Specifications |
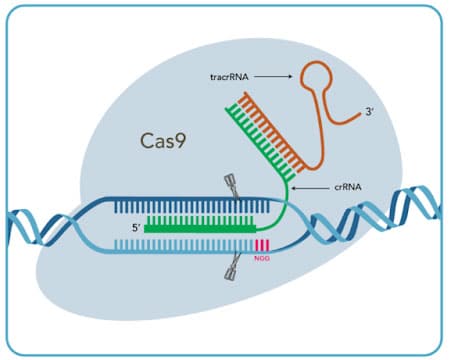
Cas9 System
 |
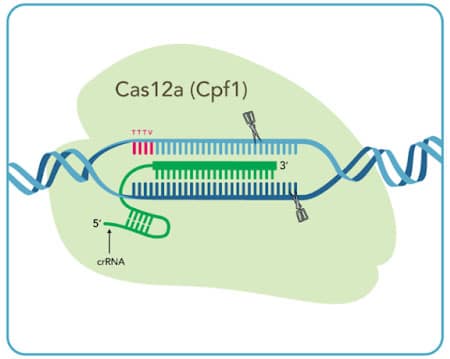
Cas12a System
 |
|---|---|---|
| Applications | General Genome Editing |
|
| Ribonucleoprotein components |
|
|
| Variants |
|
|
| Cas9 crRNA:tracrRNA (option 1) |
crRNA
tracrRNA
|
__ |
| Cas9 sgRNA (option 2) |
|
__ |
| Cas12a crRNA | __ |
|
| CRISPR enzyme |
|
|
| DNA cleavage |
|
|
| PAM sequence† |
|
|
| Current recommendations for Alt-R RNP delivery |
|
|
IDT does not sell gene therapy kits, and nothing sold by IDT should be construed as a gene therapy kit. Customers should not use any IDT products for self-administration.
CGMP gRNA products are manufactured in accordance with ICH Q7, 21CFR210, 211, and parts of 600. IDT engineering runs and CGMP gRNA are for development and investigational use only. The performance characteristics of this product have not been established. This product is not intended to be used as final drug product. The purchaser is solely responsible for all decisions regarding the intended use of the product and any associated legal or regulatory obligations.
RUO24-3155_001