IDT is proud to present a webinar on CRISPR GUARD presented by Dr. Matt Coelho. For more information on this special presentation, click here.
Background
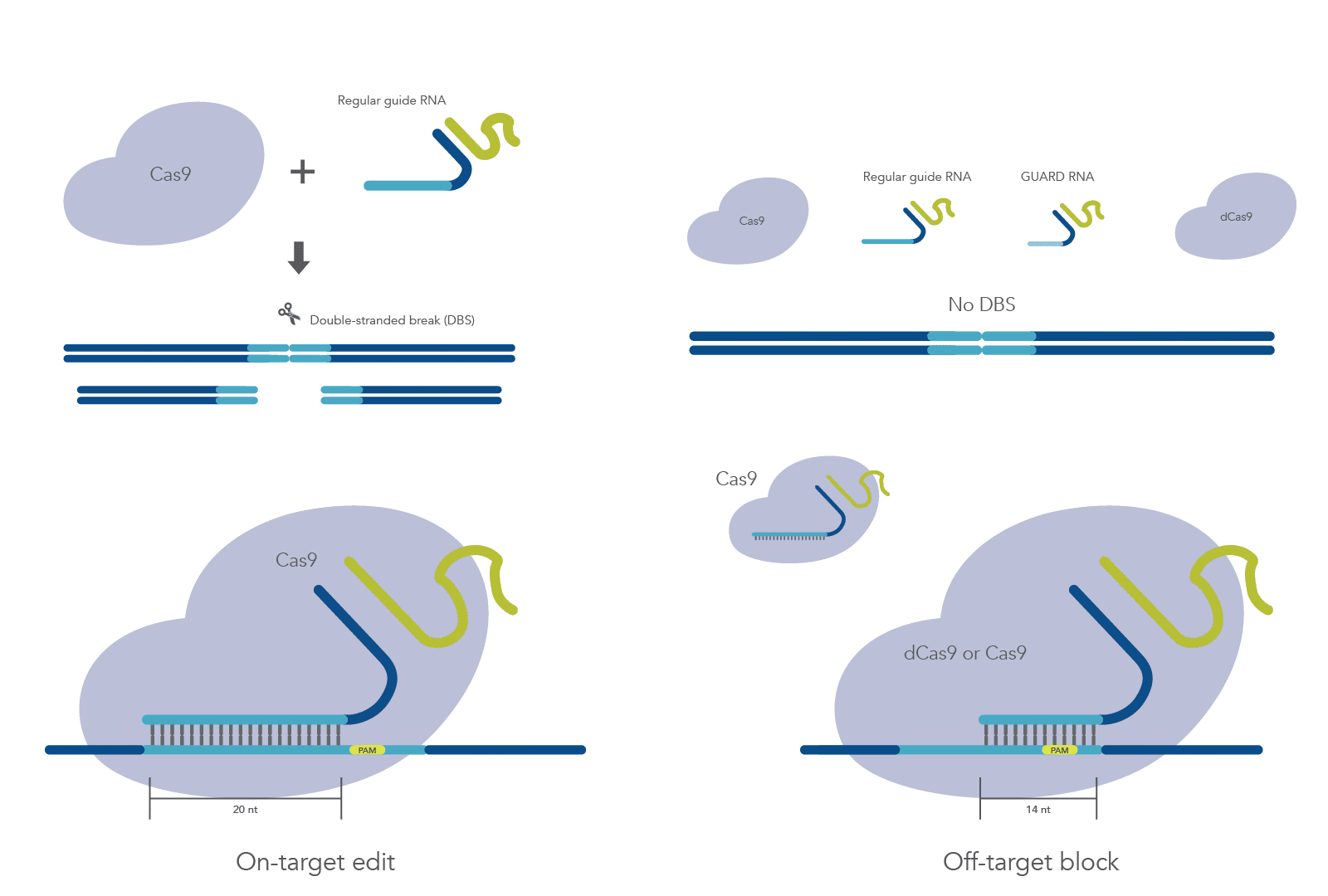
CRISPR-Cas9 systems have changed the landscape of genome editing; however, off-target Cas9 mutagenesis can elicit unwanted effects. Although gRNA design is continually improving, off-target effects cannot be eliminated when targeted editing location is short, as is the case with correction of disease-causing mutations [1,2].
A system that further reduces or eliminates off-target mutations while preserving on-target activity is needed. Previous studies have shown that gRNAs shorter than 16 nt can recruit Cas9 to the correct site, but they don’t permit nuclease activity [3–5]. Coelho et al. describe the development of a system, called CRISPR Guide RNA Assisted Reduction of Damage (CRISPR GUARD), which aims to block mismatched gRNAs from binding off-target sites through competition with an inactive Cas9 complex containing the shortened guides. They use a combination of in vitro and cellular studies to test an array of GUARD RNAs and determine the best design for blocking off-target mutations while maximizing on-target editing.
Methods
To reduce CRISPR off-target effects while also permitting on-target editing, Coelho, et al. demonstrates that co-delivery of short off-target gRNA and on-target gRNA reduce off-target editing. The feasibility of GUARD RNA competition at off-target loci was demonstrated by using a perfectly complementary 15 nt GUARD RNA against an immobilized off-target DNA template and a gRNA targeting VEGFA with four mismatches. Off-target association kinetics revealed similar binding efficiencies for GUARD RNA and mismatched gRNA.
Next, the study authors designed an array of potential GUARD RNAs for in vitro and cellular tests. Three different competitive GUARD RNAs—truncated versions of on-target gRNA incorporating mismatches found at off-target sites—were developed: one 14 nt GUARD RNA, one 15 nt GUARD RNA, and one 20 nt GUARD RNA with 15 nt target complementarity and 5 nt spacer. Additionally, Coelho et al. developed non-competitive GUARD RNAs—RNAs which bind flanking regions of off-target sites by blocking the protospacer and protospacer adjacent motif (PAM) to reduce competition at the target site.
Results
CRISPR GUARD design
Initial tests revealed that only the 15 nt competitive GUARD RNA could protect off-target sites from Cas9 cutting; however, cleavage at the target site was also impaired. Because binding and cleavage are particularly rapid in vitro, Coelho et al. pre-incubated Cas9 with a 14 nt non-competitive GUARD RNA, which achieved robust protection of the off-target site. Notably, this protection disappeared 25 bp and 50 bp away from the off-target gRNA PAM, suggesting that non-competitive GUARD RNAs are effective at blocking off-target sites only if 10 or fewer base pairs away from the gRNA PAM.
In contrast to the in vitro assays, all GUARD RNAs effectively protected the off-target site without significantly interfering with on-target cutting when transfected into Cas9-expressing HEK293 cells along with VEGFA gRNA. Dosing experiments demonstrated that higher GUARD RNA concentrations further reduced off-target effects, and the study authors recommend an optimal ratio of 5:1 GUARD RNA to gRNA.
To confirm the relevance of GUARD DNA, Coelho et al. next co-delivered inactive Cas9 complexed with an off-target specific 14 nt GUARD RNA and wild type Cas9 complexed with on-target gRNA to HEK293 cells. The GUARD RNAs effectively protected the off-target site, and there was no redistribution of active Cas9 to other off-target sites. Further investigation of various GUARD RNAs revealed that off-target loci might be more amenable to protection by CRISPR-GUARD than other sites. For example, low GC-content lowered the protective ability of the GUARD RNA in this experiment.
Coelho et al. expanded the study by testing whether multiple off-target sites could be protected simultaneously by multiplexing three GUARD RNAs together. Multiplexing was generally successful at protecting multiple off-target sites, with non-competitive GUARD RNAs yielding the best off-target protection without also interfering with on-site editing. To further evaluate the safety of GUARD RNAs, the study authors confirmed the in vitro results in HEK93 cells, showing that none of the GUARD RNAs delivered without on-target gRNA could induce Cas9-mediated genome edits.
CRISPR base editing
Based on the results, which were achieved with indel editing, the study authors next assessed CRISPR GUARD for base editing, which is more prone to off-target effects. Using HEK293 cells, they achieved robust protection of off-target cytosines by GUARD RNAs. Similar to the results with Cas9 for indel editing, the GUARD RNA did not inhibit base editing by Base Editor 3 (BE3). However, unlike with Cas9, GUARD RNA alone could elicit a small but significant number of editing events.
Identifying Functional GUARD RNAs
Finally, Coelho et al. performed a high-throughput screen to assess 600 GUARD RNAs that are 15 nt in length together with four gRNAs selected because they target disease-causing genes. Comparing these GUARD RNAs against control non-targeting (NT) GUARD RNAs, the study authors observed effective protection against off-target indels. However, off-target base editing was significantly increased with GUARD RNAs compared to NT GUARD RNAs.
The proportion of functional GUARD RNAs increased with higher GC content, and GUARD RNAs with a higher degree of spatial overlap with the gRNA, especially at the seed and PAM region, yielded superior off-target protection. These results suggest off-target protection by CRISPR-GUARD is mediated by direct competition with gRNA and highlight important considerations for GUARD RNA design.
Conclusions
Coelho et al. demonstrate that CRISPR-GUARD is an effective mechanism for preventing off-target Cas9 effects without interfering with on-target edits and can be used to protect multiple off-target sites simultaneously (Figure 1). They describe key considerations for GUARD RNA design, including distance from the off-target PAM, GC-content, and concentration.
The study authors suggest that CRISPR GUARD could be an effective tool for therapeutic applications that require high editing efficiency and where high-fidelity Cas9 variants cannot fully suppress persistent off-target editing.
From plasmid construction to CRISPR GUARD and other genome editing experiments, IDT offers a wide variety of reagents to support your research:
IDT offers a variety of plasmid construction reagents, including:
The IDT line of Alt-R™ CRISPR genome editing products, include:
References
- Cradick, T. J., Fine, E. J., Antico, C. J. & Bao, G. CRISPR/Cas9 systems targeting β-globin and CCR5 genes have substantial off-target activity. Nucleic Acids Res. 2013; 41(20): 9584–9592.
- Dever, D. P. et al. CRISPR/Cas9 beta-globin gene targeting in human haematopoietic stem cells. Nature. 2016; 539: 384–389.
- Kiani, S. et al. Cas9 gRNA engineering for genome editing, activation and repression. Nat. Methods. 2015; 12: 1051–1054.
- Dagdas, Y. S., Chen, J. S., Sternberg, S. H., Doudna, J. A. & Yildiz, A. A conformational checkpoint between DNA binding and cleavage by CRISPR-Cas9. Sci. Adv. 2017; 3:eaao002
- Dahlman, J. E. et al. Orthogonal gene knockout and activation with a catalytically active Cas9 nuclease. Nat. Biotechnol. 2015; 33: 1159-1161
For research use only. Not for use in diagnostic procedures. Unless otherwise agreed to in writing, IDT does not intend these products to be used in clinical applications and does not warrant their fitness or suitability for any clinical diagnostic use. Purchaser is solely responsible for all decisions regarding the use of these products and any associated regulatory or legal obligations. Doc ID# RUO22-0791_001