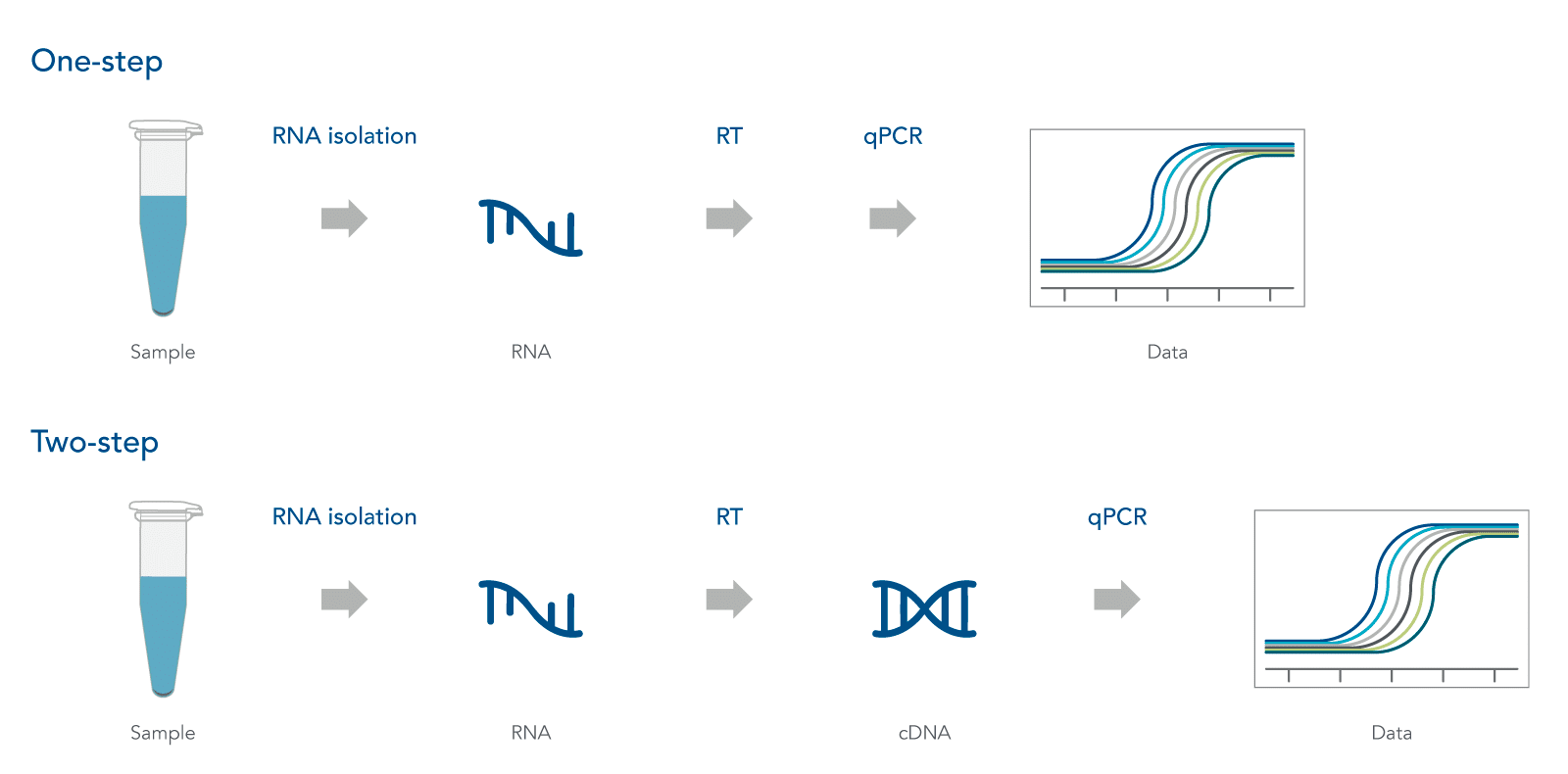
When performing real-time qPCR, one has to decide whether to use a one-step protocol that combines the RT reaction and PCR in one tube, or a two-step protocol where the RT reaction is performed separately from the PCR (Figure 1). Each method has its advantages and disadvantages that extend from the ease of logistics and cost of reaction to the resulting yield and sequence representation. One-step reactions are certainly easier to set up with less overall hands-on time, but don’t provide the flexibility and control possible with two-step reactions. One-step vs. two-step RT-PCR, which would you choose?
One-step RT-qPCR: convenience with minimal handling
In one-step RT-qPCR, reverse transcription and PCR occur in the same tube. Gene‑specific primers are used for generating the cDNA and for subsequent amplification. Major advantages of one-step reactions include limited sample handling and reduced bench time, which helps to decrease chances for pipetting errors and cross contamination. This method is quick to set up and makes processing multiple RNA samples easy when you are amplifying only a few genes of interest. It is also ideal for situations where the same genes must be amplified repetitively, such as in high-throughput identification of a specific target using well-established reaction conditions.
However, when using this protocol, you must take several concerns into account. For example, with one-step RT‑qPCR, you are fully committed to the amount and quality of the RNA used in the reaction. With regards to RNA quality, it is important to remember that common contaminants such as fatty acids, buffer salts, phenol, or other components from sample isolation can compromise the efficiency of the RT reaction which is the most sensitive part of the procedure.
With one-step RT‑PCR, all of the RNA sample is converted to cDNA and all of the cDNA is used in the PCR. So if the copy number of your target sequence is high, signal may come up too early, or if there is not enough cDNA, you may not detect your target. If you want to adjust your template concentrations, or to amplify other genes at a later date using the one-step method, you must keep an aliquot of the original RNA sample and run the entire reaction again. Because of these factors, one-step reactions may require substantially more RNA in your initial samples if you are performing multiple amplifications. Variation between these different RT reactions can significantly complicate result interpretation.
Furthermore, the combined RT and PCR reagents must allow these reactions to proceed together in one tube. This prevents use of the most effective reagents and conditions for each individual reaction. So while all of the cDNA from the RT reaction is used in the PCR in a one-step RT‑qPCR—theoretically providing very high resolution identification—the compromised reaction conditions may affect efficiency and, thus, yield [1].
The specialized systems that allow both reactions to occur together are usually supplied through commercial kits, which can be expensive. However, one-step RT-qPCR reactions are smaller and they use less reagent(s) compared to the two‑step method.
Finally, one-step RT‑qPCR requires careful evaluation to prevent primer dimer formation because the gene specific primers will be present not only during PCR cycling, but also during the lower temperature conditions of the RT reaction.
Two-step RT-qPCR: control and flexibility
In two-step RT‑qPCR, reverse transcription and PCR are performed as two separate reactions, with RNA converted to cDNA first, using a choice of random hexamers, oligo dT primers, or even gene‑specific primers. Use of random hexamers and/or oligo dT primers provides a cDNA archive of all RNA species in the sample. Either a portion of the RT reaction is diluted into the PCR, or the RT reaction can be extracted and precipitated, allowing control over the amount of cDNA input. This flexibility is an asset when working with genes of variable abundance. Any residual cDNA is available for future amplification reactions of other genes, or even for other applications. Use of the cDNA from the same RT reaction for multiple PCRs makes it easier to compare results across experiments because reaction variability and failure is limited to the PCR step, which simplifies any required troubleshooting.
Two‑step RT‑qPCR provides the opportunity for individual modification of the RT reaction and qPCR for efficiency and maximum sequence representation. These protocols typically produce higher yields of cDNA during the RT than one‑step procedures. Because of the ability to modify both reactions and the higher cDNA yields, two‑step reactions can have higher resolution than one‑step reactions. The ability to separately modify reactions is also beneficial when performing PCR on challenging sequences.
Beyond just having more control over the reactions and variability, the two-step method is particularly useful when the goal is to identify multiple messages from a single RNA sample or perform multiple PCR amplifications from a single cDNA sample.
So one-step or two-step RT-PCR?
The choice between one-step and two-step reactions comes down to the goal of your experiment. Use one-step RT‑qPCR when you have large sample quantities and/or a limited number of known targets that you may be repeatedly amplifying. It can be ideal for high-throughput identification applications that use an optimized reaction with well‑tested primers.
Use two-step RT-qPCR when your sample quantity is limited and/or you need to examine multiple targets or targets that may not be well characterized and may require you to go back to test new targets. Two‑step RT‑qPCR will also give you the control to make modifications in difficult PCR situations and when target abundance is variable.
Table 1 summarizes the required reagents, advantages, considerations, and applications for one‑step vs. two‑step RT‑qPCR protocols.
| One-step RT-qPCR protocol | Two-step RT-qPCR protocol | |
|---|---|---|
| Primers used in RT |
|
|
| Advantages |
|
|
| Considerations |
|
|
| Best for: |
|
|
References
1. Wacker MJ, Godard MP. Analysis of one-step and two-step real-time RT-PCR using SuperScript III. J Biomol Tech. 2005;16(3):266-271.
For research use only. Not for use in diagnostic procedures. Unless otherwise agreed to in writing, IDT does not intend for these products to be used in clinical applications and does not warrant their fitness or suitability for any clinical diagnostic use. Purchaser is solely responsible for all decisions regarding the use of these products and any associated regulatory or legal obligations. Doc ID: RUO23-1870_001