Blood cancer research
Blood cancer profiling has greatly benefited from next generation sequencing, but challenges remain, such as amplification of relevant complex alterations, cumbersome workflows, and bioinformatic pipeline needs. Learn about these challenges and how Archer NGS assay solutions can help.
Archer™ NGS assay solutions
Overview
Blood cancer profiling challenges with NGS
Hematology oncology research has greatly benefited from next generation sequencing (NGS) methods to identify relevant cancer-driving biomarkers. Heme malignancies have a complex biomarker spectrum, but NGS methods to detect relative fusions, splicing, expression, copy number variants, single nucleotide variants, and complex alterations have been vital to labs.
However, traditional NGS methods may not provide all relevant information and can have difficult workflows and analysis. One specific challenge in blood cancer profiling is myeloid-relevant genes rich in GC-content, such as CEBPA, are difficult to amplify and sequence with NGS. Additionally, internal tandem duplications (ITDs), such as FLT3-ITD relevant in AML, can be difficult to detect with NGS due to variable locations and lengths along with random sequence insertions which can confound genome aligners. Due to this, most NGS methods have difficulty detecting ITDs of all sizes and insertion points, and sensitivity dramatically reduces above 100 bp ITDs. Finally, there are a multitude of relevant fusions in hematological neoplasms, including many novel fusions. In fact, one study found that 41% of acute myeloid leukemias and 88% of myelodysplastic syndromes had novel fusions [1]. However, since these are relatively rare, traditional opposing primer-based NGS methods may not be designed with primers to detect all of them, thus missing potentially relevant biomarkers (Figure 1).
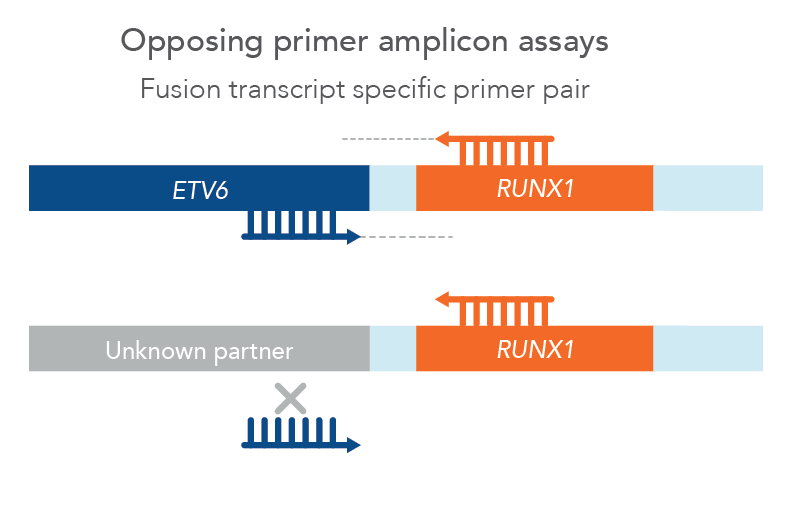
Figure 1. Opposing primer amplicon assays lack detection of unknown fusions. In this example, a primer pair for ETV6::RUNX1 can only amplify that specific fusion since the ETV6 primer cannot bind to an unknown partner, therefore missing a novel RUNX1 fusion.
Additionally, NGS library preparation workflows can be cumbersome, increasing the opportunity for manual errors. In-house bioinformatics pipelines can be expensive and time-consuming to build and maintain. They also may lack the QC and statistical tools necessary to reduce errors and guide calling, leaving it up to lab staff to make difficult calls.
Archer NGS assay solutions for blood cancer research resolve challenges with patented AMP™ chemistry, streamlined parallel workflows, and user-friendly analysis. Assays are designed to fit your lab’s unique needs whether you are looking for comprehensive blood cancer profiling with a high-throughput workflow, to small footprint assays with low-throughput workflows.
Detect more with AMP chemistry
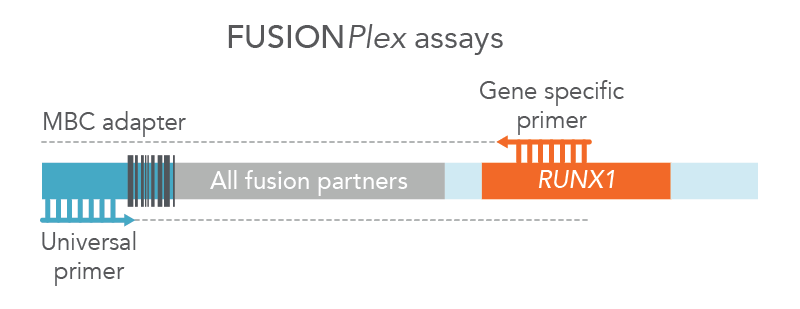
For blood cancer profiling, Anchored Multiplex PCR (AMP) is a target enrichment method that uses molecular barcoded adapters and single, nested, gene-specific primers for amplification, permitting open-ended capture of DNA, RNA, and cfDNA fragments. This approach enables the detection of novel fusions (Figure 2), allows for flexible primer design, and protects against dropout of primers. MBC adapter ligation prior to PCR combined with Archer Analysis software power the correction of systematic errors and quantitative analysis of the original unamplified template.
Figure 2. FUSIONPlex™ assays detect both known and novel fusions. FUSIONPlex assays include MBC adapters with a universal priming site. Since it is paired with the relevant gene-specific primer (in this case, RUNX1), both known and novel fusions are amplified.
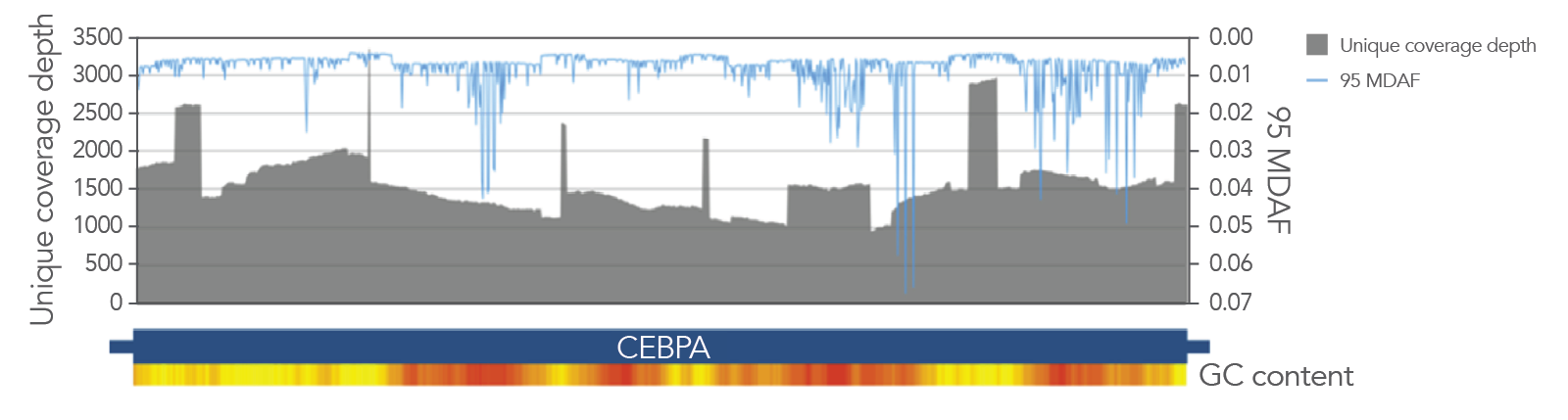
Archer blood cancer assays amplify through GC-rich regions, such as those in CEBPA, with high target coverage. Statistically significant and confident variant detection is powered by Archer’s unique outlier identification algorithm informed by normalized data to characterize position specific noise stemming from PCR and sequencing.
Figure 3. Excellent coverage of CEBPA with VARIANTPlex™ Core Myeloid panel, with 95 MDAF plotted overall. Greater than 1000X unique molecule coverage depth from 50 ng DNA input and 2 M reads with the Core Myeloid panel (gray area), the minimal allele fraction that can be differentiated from background noise 95% of the time (95 MDAF) for the least sensitive base substitution, in blue line.
VARIANTPlex blood cancer assays also provide comprehensive detection of ITDs. AMP chemistry enables tiling across both strands of DNA with primers that amplify independently of each. When reads from these primers are analyzed with the Archer Analysis pipeline, long consensus reads are generated based on sequence similarity and aligned against the genome to identify duplication events, allowing for confident detection of ITDs, including FLT3-ITD (Table 1).
Table 1. FLT3-ITDs called at ultralow allele frequencies using the VARIANTPlex Core Myeloid panel.
A FLT3-ITD-positive cell line was titrated into a background control sample. Triplicate libraries were prepared using 200 ng total input DNA and the VARIANTPlex Core Myeloid panel. ITD calling was performed using Archer Analysis.
| FLT3-ITD fraction | Input ITD molecules* | Expected ITD called |
|---|---|---|
| 0.0 | 0 | 0/3 |
| 0.75 | 45000 | 3/3 |
| 0.075 | 4500 | 3/3 |
| 0.0075 | 450 | 3/3 |
| 0.00075 | 45 | 3/3 |
| 0.00025 | 15 | 2/3 |
*Assuming 300 haploid copies per ng.
Blood cancer panels and reagent formats fit for your lab’s needs
| Genes targeted | Genomic alterations detected | Recommended reads | ||
|---|---|---|---|---|
| DNA input | VARIANTPlex | |||
| Myeloid | 75 | SNVs, indels, ITDs | 4 M | |
| Core Myeloid | 37 | SNVs, indels, ITDs | 3 M | |
| MPN Focus | 12 | SNVs, indels, ITDs | 800 K | |
| AML Focus | 11 | SNVs, indels, ITDs | 800 K | |
| RNA input | FUSIONPlex | |||
| Pan-Heme | 199 | Fusions, SNVs, indels, expression levels | 4.5 M | |
| Heme v2 | 87 | Fusions, SNVs, indels, expression levels | 1.5 M | |
| Myeloid | 84 | Fusions, SNVs, indels, expression levels | 1.5 M | |
| Lymphoma | 125 | Fusions, SNVs, indels, expression levels | 2 M | |
| Acute Lymphoblastic Leukemia (ALL) | 81 | Fusions, SNVs, indels, expression levels | 1.5 M | |
In addition to detection of the above alterations, Archer IMMUNOVerse™ assays can provide T- and B-cell clonality, minimal residual disease (MRD), and somatic hypermutation information relevant for heme malignancies. IMMUNOVerse panels have parallel workflows with VARIANTPlex and FUSIONPlex assays, allowing for comprehensive, streamlined blood cancer profiling.
Pair DNA- and RNA-based assays to efficiently generate a complete biomarker profile
| Paired panels | Genes | Reads | Samples* per NextSeq 500/550 high output kits v2.5 |
|---|---|---|---|
| FUSIONPlex Pan-Heme | 199 | 4.5 M | 47 |
| VARIANTPlex Myeloid | 75 | 4 M | |
| FUSIONPlex Myeloid | 84 | 1.5 M | 88 |
| VARIANTPlex Core Myeloid | 37 | 3 M |
Data assumes Archer libraries are prepared using the liquid reagent workflow.
*Sample = 1 VARIANTPlex library + 1 FUSIONPlex library
VARIANTPlex and FUSIONPlex panels can be paired for cost-effective solutions to provide SNVs, indels, ITDs, fusions, and expression alterations from a single sample.
Choice of assay reagent formats are available to suit your lab’s sample volumes.
- For high-throughput labs, liquid reagents are ideal for automated liquid handling workflows. Liquid adapters are available in a 96‑well plate format and include an increased number of dual indexes (over 36,000 unique combinations of sequencing indices) to support high-throughput sequencing.
- For manual workflow labs, single‑use, reaction‑size lyophilized reagents allow your lab to avoid potential contamination during manual pipetting and provide extended shelf life.
Both reagent formats have demonstrated excellent performance, with detection of 100% of expected variants in reference materials*. Regardless of reagent format, all reagents are color‑coded to match protocol steps, making benchwork easy (Figure 4).
*Data available upon request
Figure 4. Reagents are color-matched to protocol steps for easy benchwork. Each step of the protocol has a different color, which matches the packaging of the reagents (in this case, liquid reagents) for that step.
Customize your panel content
Relevant oncology biomarkers are emerging rapidly, necessitating labs to modify their assays to meet the pace of discovery. With the foundation of proprietary AMP chemistry and Archer Analysis you can confidently add targets to existing Archer assays or start from scratch without compromising assay performance. To get started, visit Assay Marketplace, the Archer assay design tool.
Blood cancer research assays
Comprehensive genomic profiling
Targeted pan-heme and myeloid malignancy panels
Specific heme malignancy panels
Clonalities, MRD, somatic hypermutation panels
Ready to start?
Talk with our technical sales team. Learn how Blood Cancer Research assays can identify key genomic alterations for your research.
Request a consultationResources
References
- Stengel A, Shahswar R, Haferlach T, et al. Whole transcriptiome detects a large number of novel fusion transcripts in patients with AML and MDS. Blood Adv. 2020. 4(21): 5393-5401.


 Processing
Processing