Whole transcriptome sequencing
In today’s competitive genomics landscape, RNA sequencing will help you quickly obtain your desired results. Our xGen RNA sequencing product offerings cover a wide range of diverse inputs while maintaining predictable outputs, efficiencies, and coverage metrics.
xGen™ NGS—made to differentiate.
Overview
- Fast and cost-efficient workflow—curated for RNA sequencing
- Flexible—prepare RNA-seq libraries from a wide range of sample types and input quantities
- Powered by Adaptase™ technology—for adapter tailing and ligation to 1st strand DNA; does not require 2nd strand DNA and leads to stranded RNA libraries
- Customizable—easily combine a hybridization capture workflow for targeted RNA-seq
- Combine the xGen Normalase™ Module with your RNA-seq workflow—for fast and easy library balance and pooling
- Automation-friendly RNA-seq workflow
What is whole transcriptome sequencing?
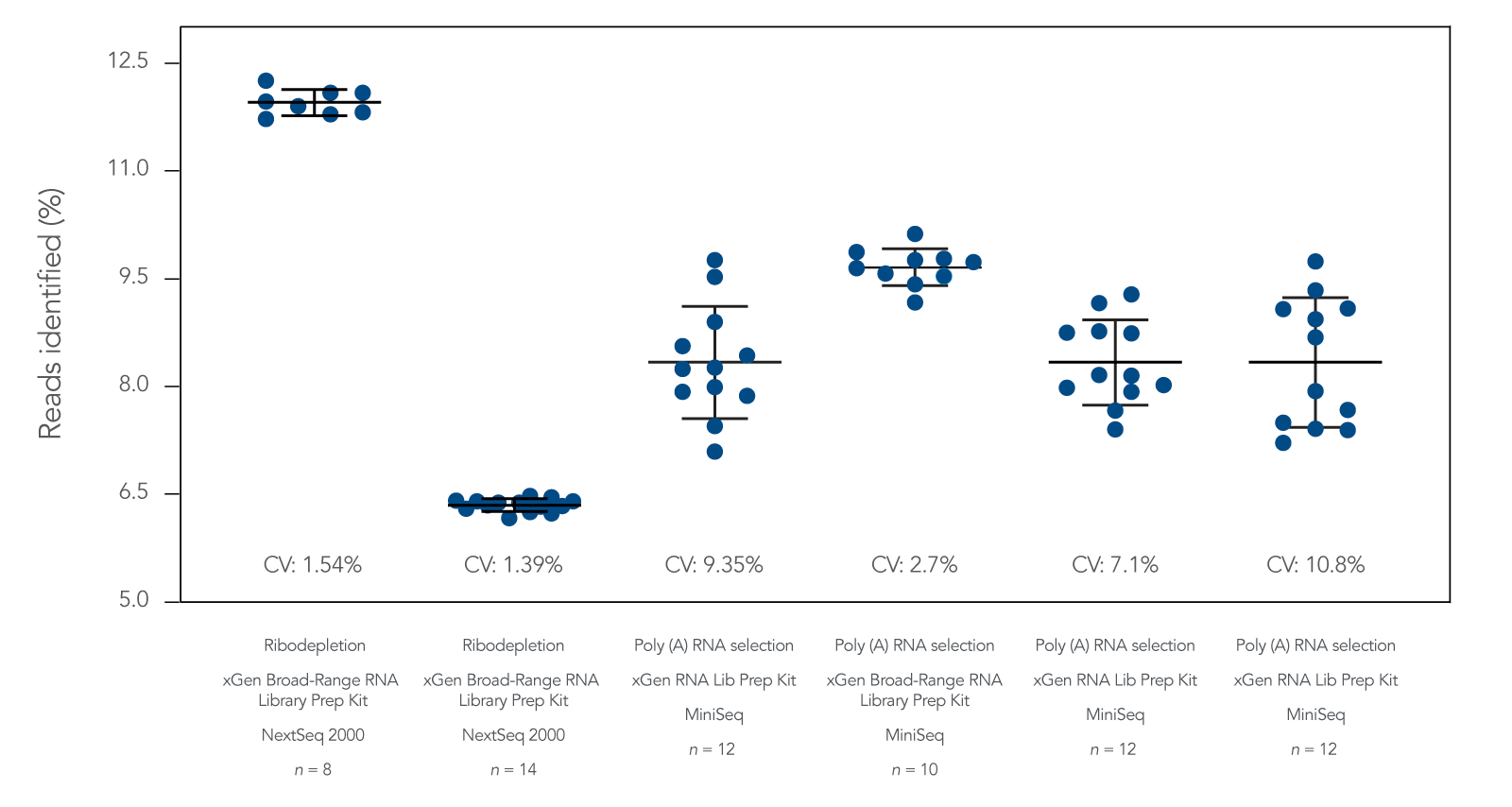
RNA sequencing (RNA-seq) is a next generation sequencing method for analyzing the transcriptome of cells and assessing gene expression variation. The xGen RNA Library Prep Kit and the xGen Broad-Range RNA Library Prep Kit are fast transcriptomics research workflow solutions with comprehensive transcript coverage and NGS data quality.
The workflow used in these kits employs Adaptase™ technology which adds a single-stranded R2 Stubby Adapter onto the 3’ end of the 1st strand cDNA product. This technology does not require 2nd strand cDNA synthesis and degradation and results in stranded RNA libraries. While the xGen Broad-Range RNA Library Prep Kit supports a wider range of total RNA inputs (from 10 ng to 1 µg) than the xGen RNA Library Prep Kit (from 100 ng to 1 µg), the latter offers a faster workflow with fewer steps. This RNA-seq workflow is compatible with upstream mRNA enrichment and rRNA depletion modules. It is easily combined with a hybridization capture workflow for a targeted mRNA capture sequencing study.
For high-throughput labs, the 96-reaction size group of products includes the xGen Normalase Module, a proprietary enzymatic library normalization kit for multiplexing. The method eliminates the need for individual sample quantification followed by manual equimolar pooling. Instead, the fragments are indexed by PCR using the included plate of xGen Normalase UDI Primer Pairs followed by the xGen Normalase Module. Final normalized library pools are then ready for Illumina-based multiplex sequencing.
Extraction
rRNA depletion or mRNA enrichment
Library prep
xGen RNA Library Prep Kit (everyday samples)
xGen Broad-Range RNA Library Prep Kit (low-quality or low-input samples)